Dec 2, 2024
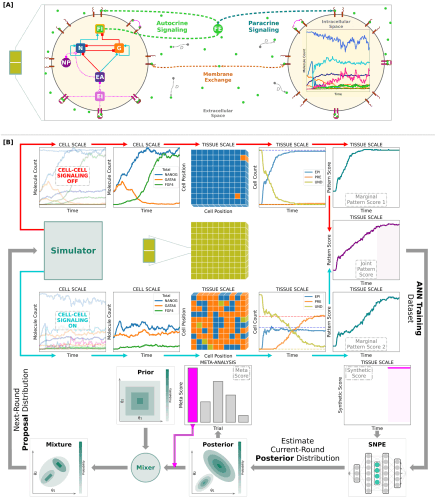
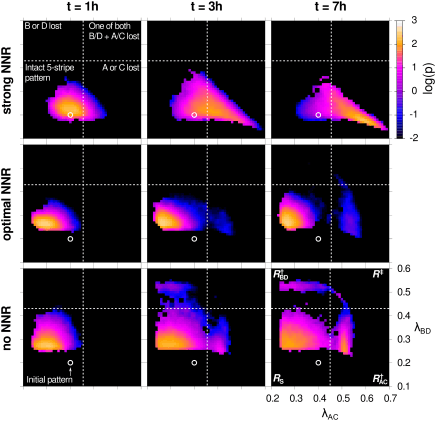
In collaboration with Maciej Majka and Marcin Zagórski (Jagiellonian University, Cracow), Nils Becker (DKFZ Heidelberg) and Pieter Rein ten Wolde (AMOLF, Amsterdam) we publish our article "Stable developmental patterns of gene expression without morphogen gradients", again in PLoS Computational Biology.